Why Reusability Is Still So Hard in Bioinformatics Pipelines
Anyone who’s built pipelines in bioinformatics knows the feeling:
- You meant to containerize…
- You meant to write down which reference files and parameters you used…
- You meant to document all your runs…
But the project was due. The experiment was changing. The team was asking for results yesterday. And so you shipped the one-off. Again.
Reusability remains one of the most desirable and elusive goals in bioinformatics. It's not that we don't want it. It's that the tools, incentives, and workflows make it exceptionally difficult to achieve. And the cost of skipping it usually hits weeks or months later, when someone asks a simple question like, “Can you just re-run that analysis?”
Let’s break down why reusability is hard, really hard, and what today’s bioinformatics toolkit actually offers in response.
The Five Fracture Points of Reusability
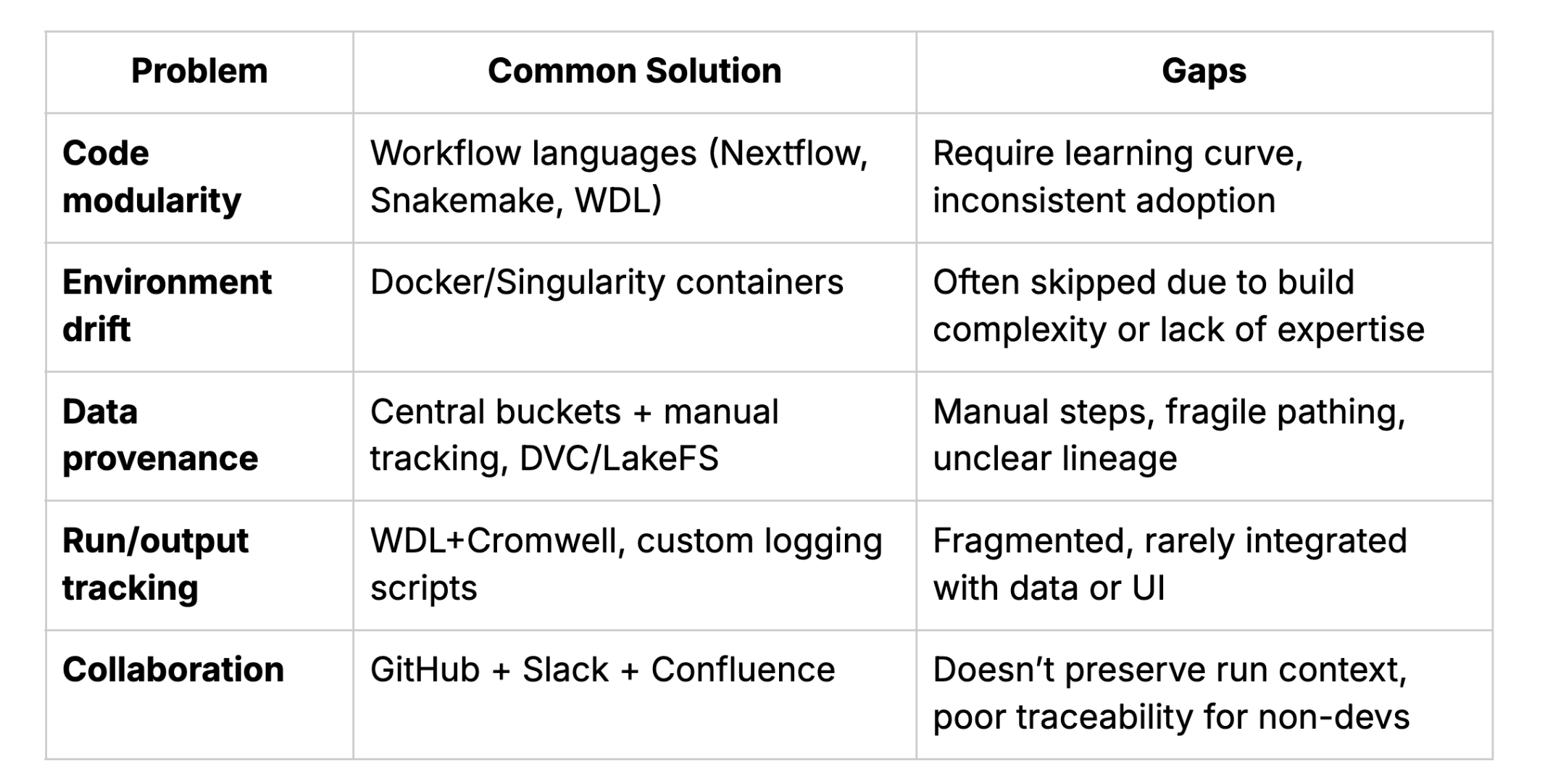
1. Code Modularity
Most pipelines are written as linear scripts, optimized for a single run. Inputs are hardcoded and outputs are routed wherever the author’s machine defaults to. There’s no abstraction between “what this does” and “how it’s used.”
- Why it fails: Code that isn’t modular can’t be reused. You’d have to copy-paste and manually tweak each time. That’s how analysis logic forks and drifts over time.
- What helps: Workflow engines (like Nextflow, Snakemake) encourage modularization by design. But adoption depends on team maturity and training, and that often takes a back seat to higher-priority tasks, even when the intention to train and align is present.
2. Environment Drift
You might be able to re-run the same code but will your current environment support it? Version mismatches in R, Python, Bioconductor, or even Bash utilities can derail an entire analysis.
- Why it fails: Environments change fast and tools get updated. One unpinned dependency can break everything.
- What helps: Containerization (Docker, Singularity) can preserve environments. But building and maintaining containers still requires expertise, alignment, and maintenance.
3. Data Provenance
Code is part of the story, and data is the other part. What exact file did this process run on? Was it the raw FASTQ, the trimmed version, or something someone emailed you labeled “final_FINAL”?
- Why it fails: Data paths are often implicit and scattered. Renaming or relocating files breaks reproducibility.
- What helps: Using centralized, versioned data storage (object stores with metadata layers like S3 + DVC, Quilt, or LakeFS) helps, but many teams still rely on ad hoc folder conventions.
A Note on FAIR DataData provenance is one pillar of a broader framework known as FAIR: Findable, Accessible, Interoperable, and Reusable. FAIR has become a guiding ideal for how scientific data should be managed, shared, and reused, especially in regulatory and collaborative settings.
But FAIR isn’t just about clean metadata - it requires consistent identifiers, standardized formats, and clear data lineage. And while it’s often discussed at the dataset level, the same principles apply to pipelines: if you can’t trace what data was used, how it was processed, and where it lives, reusability breaks down.
4. Run Logging and Output Tracking
Assuming that the code and data are there, you still need to know how they were combined: what parameters were used, what changed between versions, and where the outputs are.
- Why it fails: Most teams don’t log this rigorously. Outputs are stored manually. Parameters are set inline and forgotten.
- What helps: Workflow engines that support parameter tracking and audit logs (like WDL+Cromwell) help. But only if runs are centralized, not scattered across laptops and clusters.
5. Collaboration and Handoffs
Code reusability doesn’t exist in isolation. It has to survive a team change, a laptop refresh, a handoff to someone who wasn’t on the original Slack thread.
- Why it fails: Documentation is scarce. Personal context is lost. Git repos are created but never pushed.
- What helps: Git and GitHub/GitLab help manage version control but they don’t store run context, outputs, or data. Without additional systems, even “open” code can be opaque.
The Patchwork Toolkit of Today
So how are teams solving these problems now? Here’s what the current toolkit looks like:
Wrapping up
Every company has a reproducibility story. A key member left. The pipeline broke. A regulatory official asked a clarifying question, and no one could find the original data or exact run. A promising dataset sat idle because no one could get the original environment to work again.
The irony is that most teams intend to fix this. But reusability requires coordination across tools that weren’t built to talk to each other: across job schedulers, cloud storage, git, workflow managers, containers, metadata schemas, and collaboration platforms.
At Via Scientific, we develop tools and technologies that build on top of the best practices of existing solutions. We then combine them into a single system that makes this coordination easier, and more importantly, part of every team member's workflow. We believe that the companies that solve these problems will be able to move their science ahead and gain an advantage in their fields.
Reusability is a must, and it pays to make it the default and not the exception.







